Harvest Variants
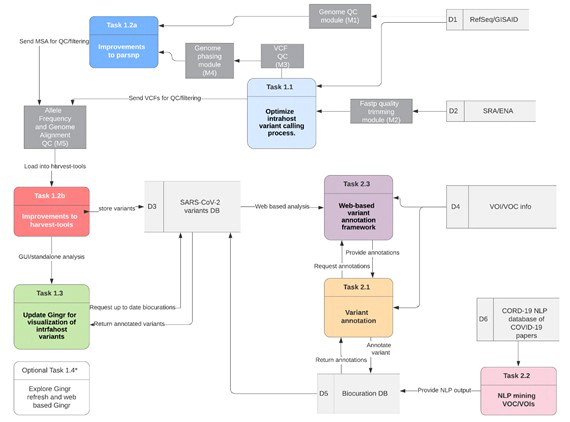
Real-time monitoring of viral mutations of circulating SARS-CoV-2 lineages, both at both the intrahost and interhost level, is a key step in understanding changes to SARS-CoV-2 infectivity/transmissibility, vaccine efficacy, and fitness within human hosts. Since the onset of the COVID-19 pandemic, there have been several variants of note in the Spike glycoprotein that have been linked to increased infectivity and are under active investigation, including A701B, D614G, E484K, K417N, N501Y, and P681H. Previous work by our group includes the Harvest package, which features three software tools: (1) Parsnp: multiple genome alignment and SNP typing, (2) Harvest tools: variant analysis file conversion and fasta data interchange format, and (3) Gingr: interactive graphical user interface for simultaneous visualization of variants, phylogeny, synteny, annotations of hundreds of thousands of genomes. The Harvest software suite, including Parsnp, harvest tools, and Gingr, were originally designed for intraspecific multiple genome alignment, variant detection, and simultaneous visualization of phylogeny and multiple sequence alignments, respectively. The Harvest suite was published 2014, and has been available on Github (https://github.com/marbl/harvest) since 2013 and supported for over 7 years. While these tools have been widely adopted by the community, they require several improvements to maximize their potential for integrated, collaborative variant tracking of SARS-CoV-2. Here we propose the development of Harvest Variants, a SARS-CoV-2 specific enhancement tio harvest tools for (i) adding support for minor variants to harvest tools, (ii) algorithmic enhancements to parsnp and harvest tools and (iii) genotype-to-phenotype tracking of curated SARS-CoV-2 variants within the Gingr graphical user interface.