 KOMB Pipeline
KOMB PipelineBackground
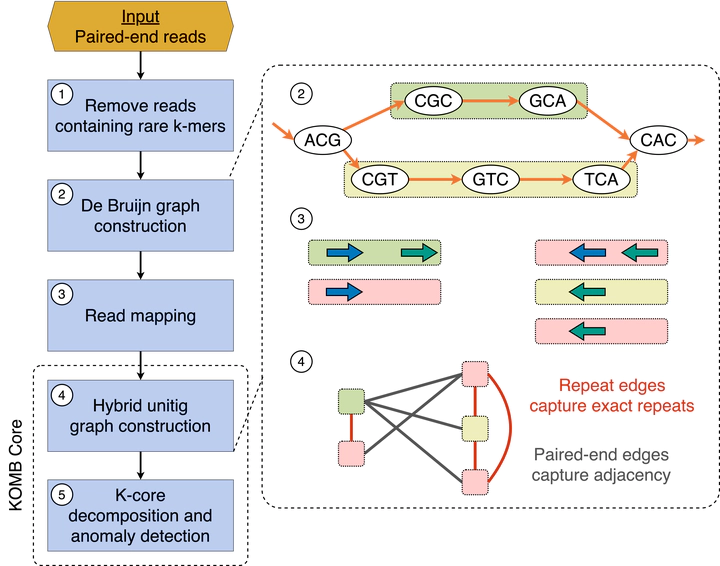
Characterizing metagenomes via kmer-based, database-dependent taxonomic classification has provided insight into underlying microbiome dynamics. However, novel approaches are needed to track community dynamics and genomic flux within metagenomes particularly in response to perturbations. We describe KOMB, a novel method for tracking genome level dynamics within microbiomes. KOMB utilizes K-core decomposition to identify flux in repetitive and homologous regions within microbiomes. K-core decomposition partitions the graph into shells containing nodes of degree at least K, yielding reduced computational complexity compared to prior approaches. Through validation on a synthetic community, we show KOMB recovers and profiles repetitive genomic regions in the sample. KOMB is shown to identify functionally-important regions in Human Microbiome Project datasets, and was used to analyze longitudinal data and identify keystone taxa in Fecal Microbiota Transplantation (FMT) samples. In summary, KOMB represents a novel graph-based reference-free approach to microbiome characterization.
Collaborators
- Dr. Santiago Segarra (Rice University)
- Dr. Charlie Seto (NCBI)
- Dr. Tor Savidge (Texas Children’s Hospital)