DYNAMITE
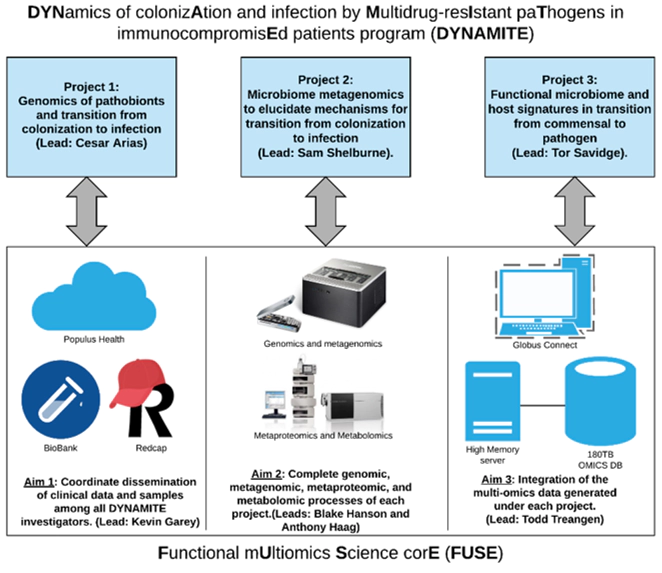 Photo by rawpixel on Unsplash
Photo by rawpixel on UnsplashProject leads
- Dr. Cesar Arias (Houston Methodist)
- Dr. Tor Savidge (Texas Childrens)
- Dr. Sam Shelburne (MD Anderson)
Summary
Antimicrobial resistance (AMR) in community and hospital-associated pathogens has been named one of the most pressing public health priorities by the United Nations. Among the most relevant multidrug-resistant (MDR) bacteria, vancomycin-resistant enterococci (VRE), extended spectrum ?-lactamase producing/carbapenem-resistant Enterobacteriaceae (ESBL-E/CRE) and Clostridiodes difficile are considered high priority inasmuch as these organisms commonly infect severely ill and immunocompromised patients, and there is a paucity of therapeutic options to treat infections caused by these bacteria. For each of these key pathogens, the intestines are the site of initial colonization and, under the influence of broad- spectrum antimicrobial therapies, these organisms can ?dominate? the gastrointestinal tract increasing the risk of clinical disease. Importantly, it is becoming progressively clear that colonization of the intestines by either VRE, ESBL-E/CRE, or C. difficile is markedly associated with subsequent colonization by other members of this group, but whether pathogen-to-pathogen signaling plays a role is not known. Further, data generated from microbiome-based studies to date has not allowed for clinically impactful interventions due to the imprecise identification of high-risk patients and it is currently unclear why only a subset of patients, under apparently similar conditions, develop colonization/disease. Our overarching hypothesis is that patient susceptibility to gut-derived nosocomial colonization and subsequent infection is critically dependent on functional microbiota- pathogen interactions that can be detected via a holistic combination of pathogen, host, and commensal microbiota analyses. The DYNAMITE program (Dynamics of Colonization and Infection by Multidrug-Resistant Pathogens in Immunocompromised and Critically Ill Patients) seeks to fill these important gaps in knowledge. Indeed, we have identified keystone microbiota features that are broadly protective against gut-derived pathogens via previously unappreciated antimicrobial mechanisms suggesting that lack of such organisms may be a critical factor in determining pathogen colonization and infection.
The aims of the program are:
- Dissect the main microbial, clinical and antimicrobial resistance determinants that impact colonization and infection by VRE, ESBL-E/CRE and C. difficile
- Evaluate the role of the commensal microbiota in VRE, ESBL-E/CRE and C. difficile colonization
- Define the functional aspects of keystone microbiota and mechanisms of protection against colonization/infection.