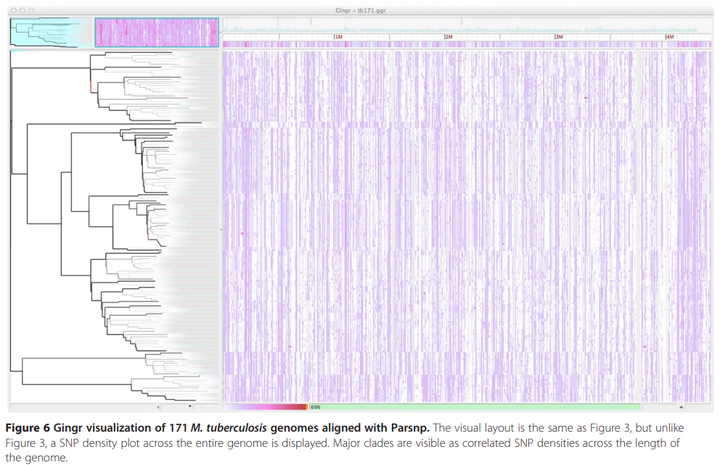
 Gingr visualization of 171 M. tuberculosis genomes aligned with Parsnp. Major clades are visible as correlated SNP densities across the length of the genome.
Gingr visualization of 171 M. tuberculosis genomes aligned with Parsnp. Major clades are visible as correlated SNP densities across the length of the genome.Background
Whole-genome sequences are now available for many microbial species and clades, however existing whole-genome alignment methods are limited in their ability to perform sequence comparisons of hundreds of sequences simultaneously. Parsnp is a fast core-genome multiple genome aligner that is able to efficiently align thousands of closely related microbial sequences. By only looking at the core-genome, Parsnp can accurately identify core SNPs across all sequences, enabling the reconstruction of large phylogenies of closely related species. Released in 2014, Parsnp is still actively used by the community and maintained by the Treangen lab.
Collaborators
- Dr. Brian Ondov (NIH)
- Dr. Sergey Koren (NIH)
- Dr. Adam Phillipy (NIH)