Background
We developed PlasmidHawk, a linear time pan-genome alignment-based pipeline to predict the lab-of-origin of unknown sequences. Compared to the previous deep learning method, PlasmidHawk has higher prediction accuracy. PlasmidHawk can successfully predict unknown sequences’ depositing labs 76% of the time and 85% of the time the correct lab is in the top 10 candidates. In addition, PlasmidHawk can precisely single out the signature sub-sequences that are responsible for the lab-of-origin detection. PlasmidHawk represents an explainable and accurate tool for lab-of-origin prediction of synthetic plasmid sequences

PhD student from September 2018 through January 2022 (currently Sr. Bioinformatics Scientist at Illumina)
Dr. Wang is a Bioinformatics Scientist at Illumina, and finished her PhD in the Treangen Lab December 2021. Previously, Dr. Wang obtained B.S. degrees in Biotechnology from Hong Kong Baptist University and MS in Biotechnology from Northwestern University. During her undergraduate, she did research in University of Chinese Academy of Sciences, Beijing University of Chemical Technology and Capital Medical University, focusing on using bioinformatics and experimental approaches to solve various life science problems, including synthetic biology, developmental biology, oncology and drug discovery. Her interest is to improve human health and environment by understanding complex biology data.

Postdoctoral Scientist from August 2019 through April 2022
Leo (NLM Postdoctoral Fellow, primary mentor Prof. Lauren Stadler, secondary mentor Prof. Todd Treangen) received his PhD in Computer Science at Rice University in 2019 working on statistical modeling of DNA sequence evolution. He was advised by Dr. Luay Nakhleh, the J.S. Abercrombie Professor and Chair of the Department of Computer Science at Rice. Since joining at Rice, Leo was awarded a graduate research fellowship from the National Library of Medicine, has published work in computational biology in journals such as Bioinformatics, presented research at scientific conferences like RECOMB-CG in Barcelona and WABI in Helsinki, and contributed to a soon to be released book on computational modeling of evolutionary histories of genomes.
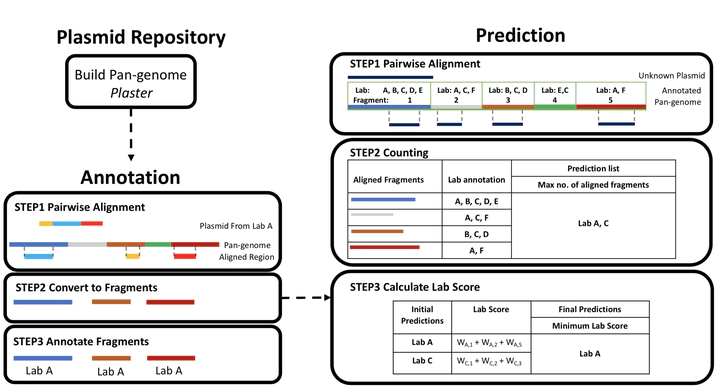 Plasmidhawk pipeline
Plasmidhawk pipeline