 Variabel Algorithm and Pipeline
Variabel Algorithm and PipelineBackground
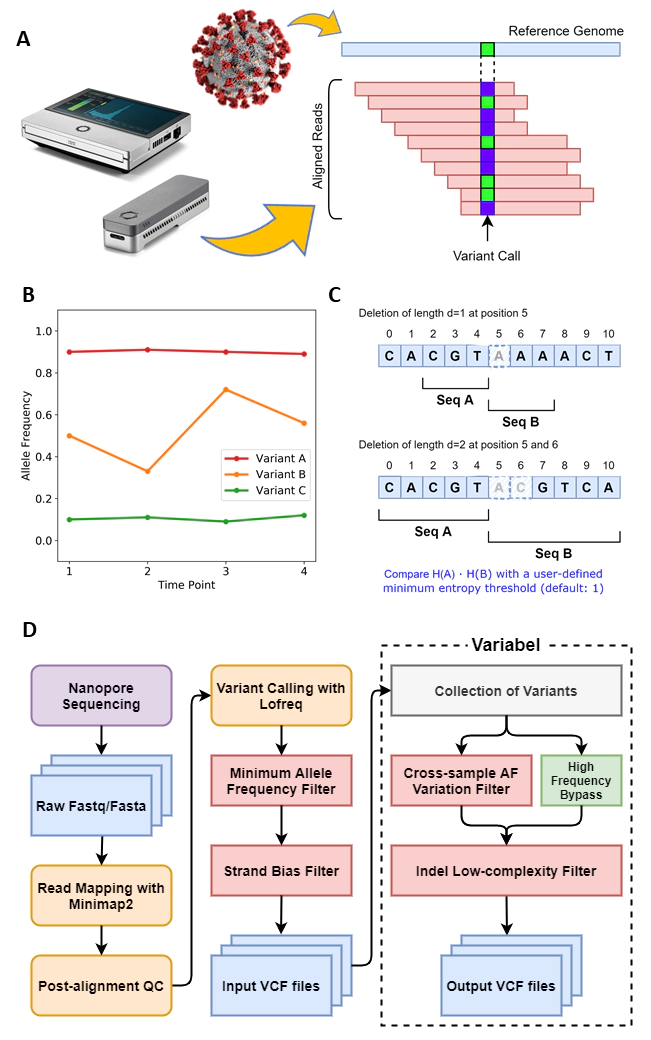
Infectious disease monitoring on Oxford Nanopore Technologies (ONT) platforms offers rapid turnaround times and low cost, exemplified by well over a half of million ONT SARS-COV-2 datasets. Tracking low frequency intra-host variants has provided important insights with respect to elucidating within host viral population dynamics and transmission. However, given the higher error rate of ONT, accurate identification of intra-host variants with low allele frequencies remains an open challenge with no viable computational solutions available.
Findings
In response to this need, we present Variabel, a novel variant call filtering tool that is able to recover intra-host variants from ONT data alone, for the first time, by exploiting the tendency of true variants to change in allele frequency across samples. We evaluated Variabel on both synthetic data (SARS-CoV-2) and patient derived datasets (Ebolavirus, Norovirus, SARS-CoV-2); our results show that Variabel can accurately identify low frequency variants below 0.5 allele frequency, outperforming existing state-of-the-art ONT variant callers for this task.
Conclusions
Vulcan is the first method designed for rescuing low frequency intra-host variants from ONT data alone. Variabel is open-source and available for download at: www.gitlab.com/treangenlab/variabel.
Collaborators
- Dr. Fritz Sedlazeck (BCM HGSC)
- Dr. Medhat Mahmoud (BCM HGSC)