[TOC]
Bakdrive: Identifying the Minimum Set of Bacterial Driver Species across Multiple Microbial Communities
The impact of microbiota on human health
Microbes form a complex and dynamic system
Disruption of microbial communities observed in many diseases,
e.g. recurrent Clostridioides difficile infection (rCDI),
inflammatory bowel disease (IBD)
Popular treatment for dysbiosis
e.g., fecal microbiota transplantation (FMT), probiotics
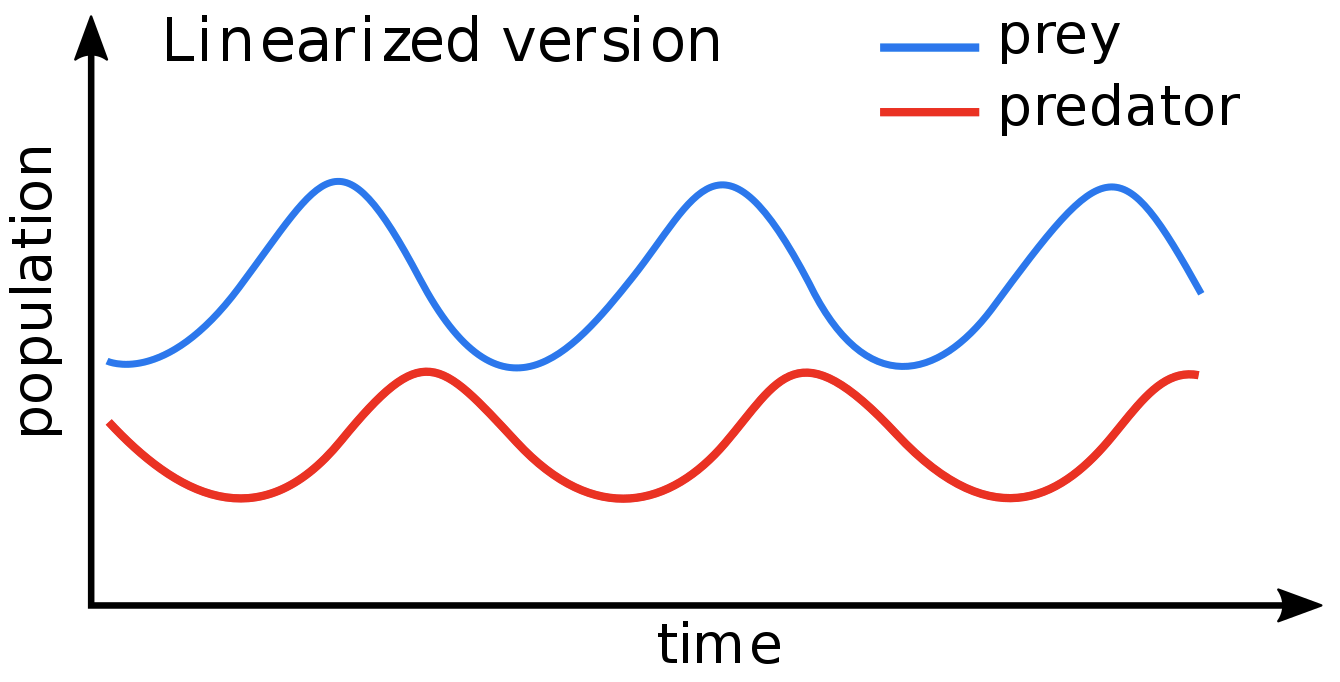
Exploring microbial communities with control theory
Control theory : a discipline of design strategies aimed at controlling dynamical systems
- e.g. system characterization, control optimization etc.
Desired Target State
Characterizing metagenomic states with microbial abundance profiles
Metagenomic states are characterized by abundance profiles .
Abundance Profiles
Angulo, Moog, and Liu, “A Theoretical Framework for Controlling Complex Microbial Communities.” (2019)
Characterizing underlying mechanisms of microbial communities
Ecological network (bacterial interaction network)
Node: species x
Edge: interspecies interaction
The abundance of each species changes following the GeneralizedLotka-Voltera(GLV) model
Lotka-Voltera model
Goal & Challenges
Goal
Find the minimum number of driver species (driver nodes), whose control can shift microbial communities from diseased to healthy states
Challenges
How to infer bacterial interactions?
How to find driver species?
Different metagenomic samples have different ecological networks. How to find a common set of driver species that works for multiple metagenomic samples ?
Flux balance analysis (FBA) is utilized to infer bacterial interactions
How to infer bacterial interactions?
FBA: Calculate the flow of metabolites through genome-scale metabolic models, thereby to predict the growth rates of organisms
Competitive (negative): consume same resources
Cooperative (positive): consume metabolites produced by another species
Software: MICOM
Christian Diener, Sean M. Gibbons, and OsbaldoResendis-Antonio, “MICOM: Metagenome-Scale Modeling To Infer Metabolic Interactions in the Gut Microbiota,” MSystems 5, no. 1 (February 25, 2020),https://doi.org/10.1128/mSystems.00606-19.
Ecological Network
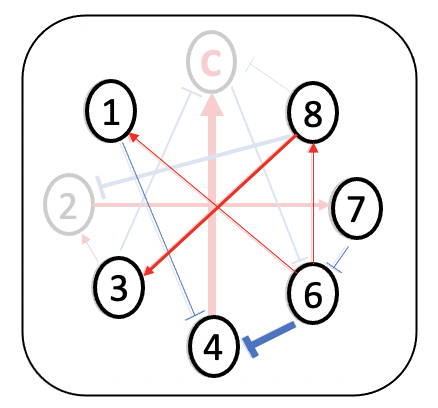
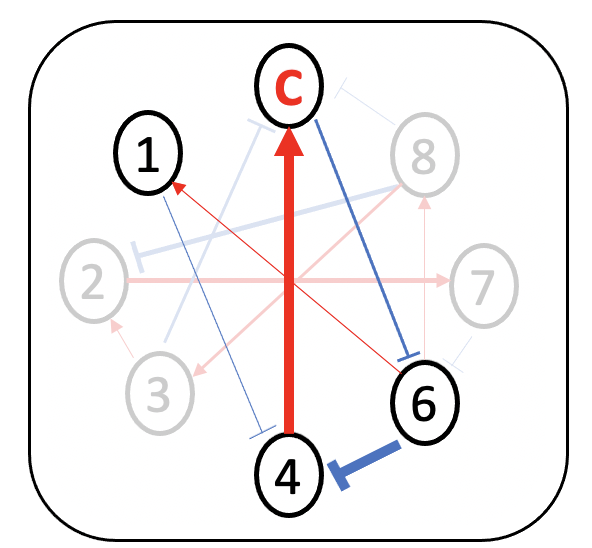
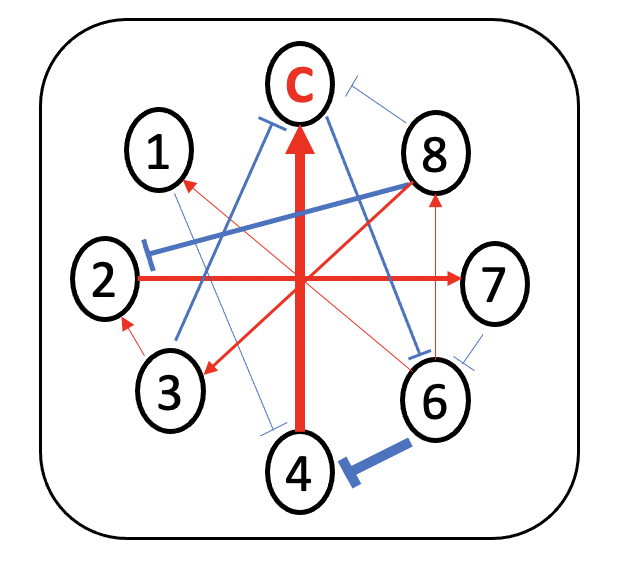
Minimum dominant set (MDS)-based approach is employed to detect driver species
How to find driver species?
- minimum dominant set (MDS) of nodes
the minimum number of nodes, which directly connect all the other nodes in the network.
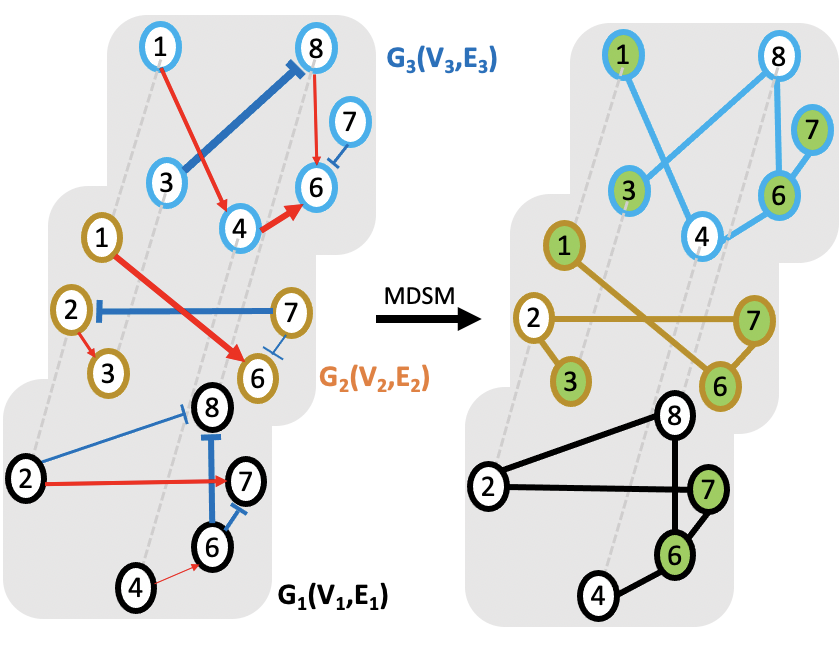
A set of metagenomic samples are characterized as a multilayer ecological networks
Different metagenomic samples have different ecological networks. How to find a common set of driver species that works for multiple metagenomic samples?
- multilayer MDS (MDSM) algorithm
minimize no. of driver nodes
Subject to
driver nodes & their neighbors cover thewholenetwork
Jose C.Nacheret al., “Finding andAnalysingthe Minimum Set of Driver Nodes Required to Control Multilayer Networks,” Scientific Reports 9, no. 1 (January 24, 2019): 576,https://doi.org/10.1038/s41598-018-37046-z.
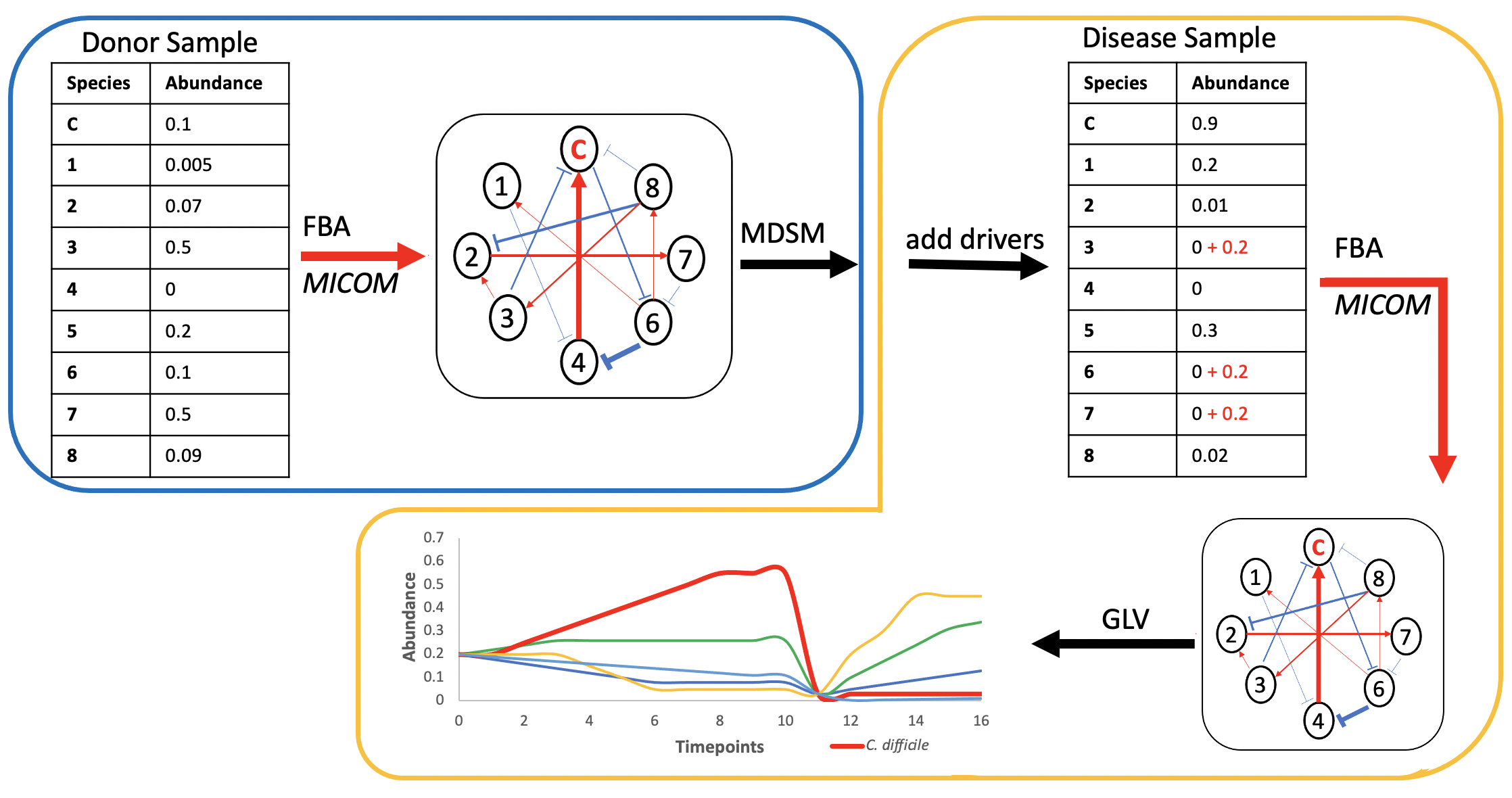
Bakdrive Pipeline
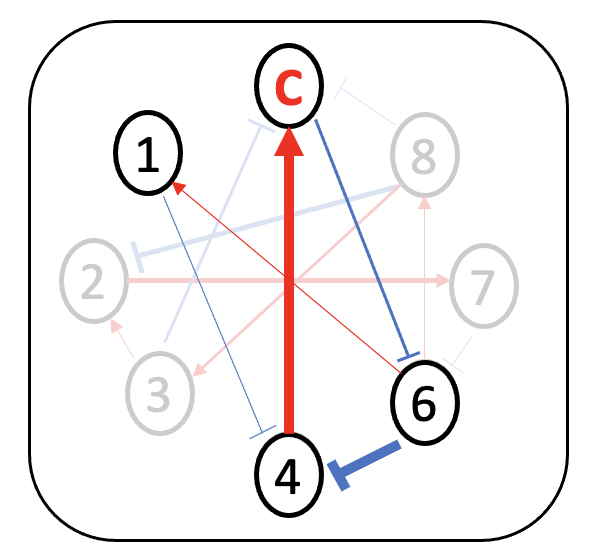
Driver species
(Probiotics)
1,3,6,7
Results Outline
Simulated Data
recurrent C. difficile infection
Known ecological networks & target species
Real Data
recurrent C. difficile infection
Unknown ecological networks & target pathogens
rCDI and FMT Simulation
*GLV: generalizedLotka–Volterra
Pool of 100 species
Select no. of species
bowel cleansing
YandongXiao et al., “An Ecological Framework to Understand the Efficacy of Fecal Microbiota Transplantation,” Nature Communications 11, no. 1 (July 3, 2020): 3329,https://doi.org/10.1038/s41467-020-17180-x.
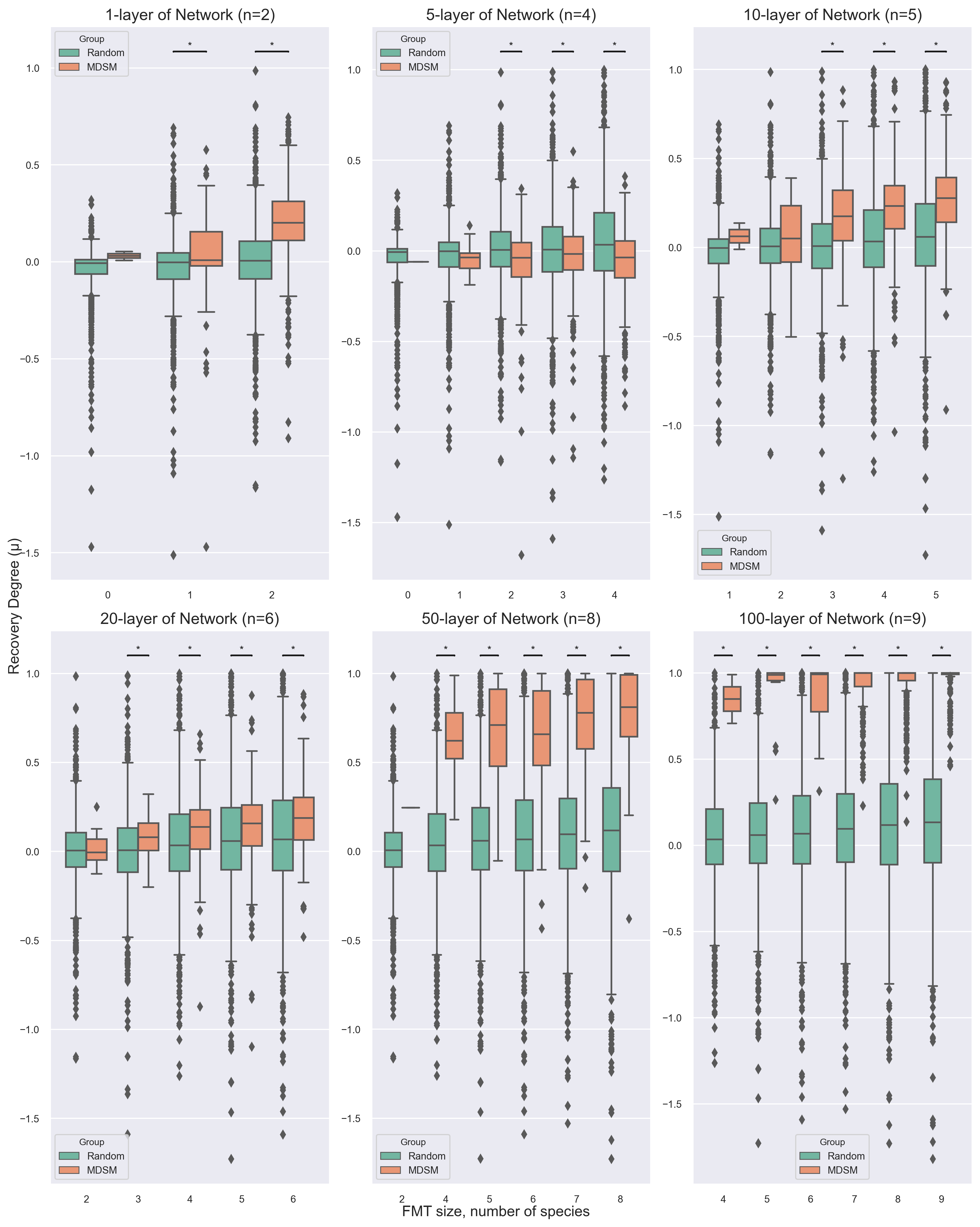
Part1 Simulated Data Case1 Universal microbial dynamics
Part1 Simulated Data Case1 Universal microbial dynamics
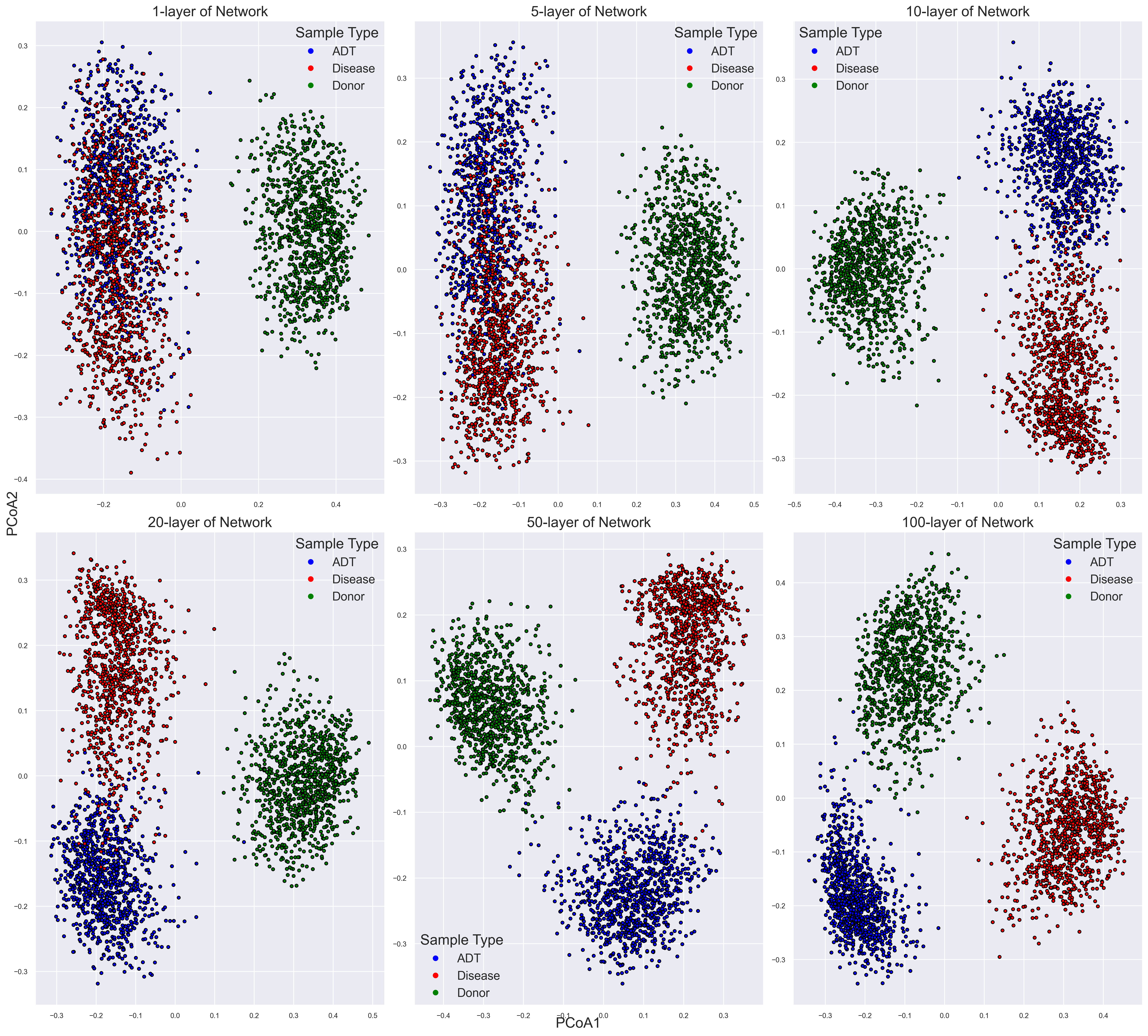
Shifts of microbial communities from diseased to healthy state
PCoAbased on Bray-Curtis dissimilarity
Part2 Real Data C. difficile infection
rCDI(n=19), donors (n=26) , FMT samples
12 patients with a single does of successful FMT
The most abundant genera:
- Klebsiella and Escherichia (Disease)
- Actinobacteria, Clostridia, Bacteroidia (Donor)
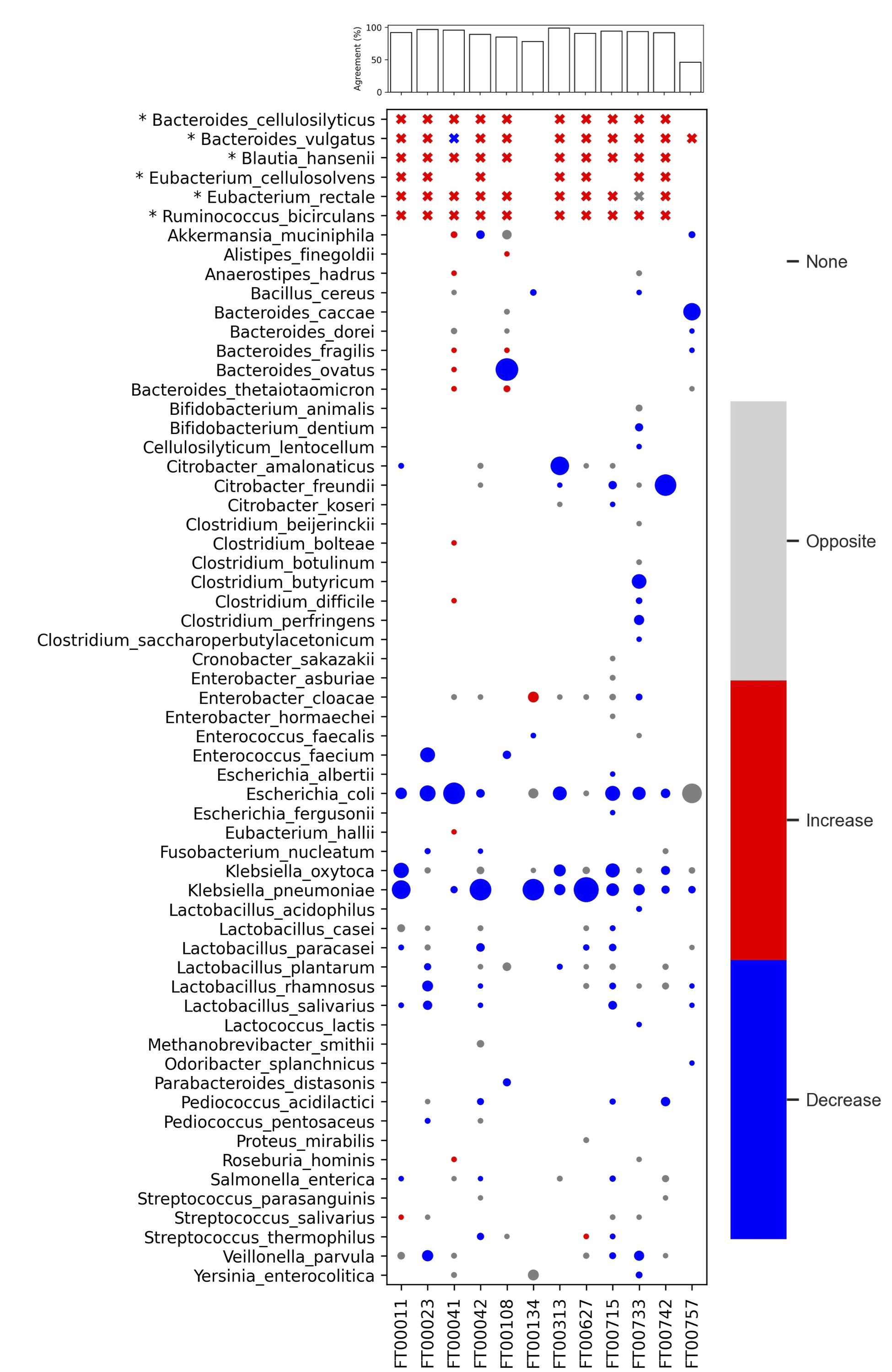
Bakdrive real data analysis
Part2 Real Data Case1 rCDI
Identify 8 driver species from 26 donor samples
Campylobacter jejuni and Streptococcus agalactiae are pathogens (removed from driver species transplantation)
Agreement = % species in therCDIsamples with the same shift between driver species transplantation (simulated data) & after-FMT (real data)
Section Summary Controlling Biological System
Limitations and Future Work:
Take the directionality and strength of ecological networks into consideration.
Apply Bakdrive in real clinical setting
Bakdrive is the first ready-to-use pipeline that detect driver species and customize probiotic cocktail
Bakdrive can directly take abundance profiles as input
Does not require known ecological networks
Does not require known target pathogens
Bakdrive takes host variations into consideration